FindPower
xiayh17
2021-07-03
Last updated: 2021-07-03
Checks: 7 0
Knit directory: wgcna-workflow/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210703) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version e65c504. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rproj.user/
Untracked files:
Untracked: LICENSE
Untracked: data/all_for_wgcna/
Untracked: data/project/
Unstaged changes:
Modified: analysis/_site.yml
Modified: data/README.md
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/FindPower.Rmd) and HTML (docs/FindPower.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | e65c504 | xiayh17 | 2021-07-03 | add findpower section for myproject |
| html | f603eba | xiayh17 | 2021-07-03 | Build site. |
| Rmd | 42955dd | xiayh17 | 2021-07-03 | Change theme for myproject |
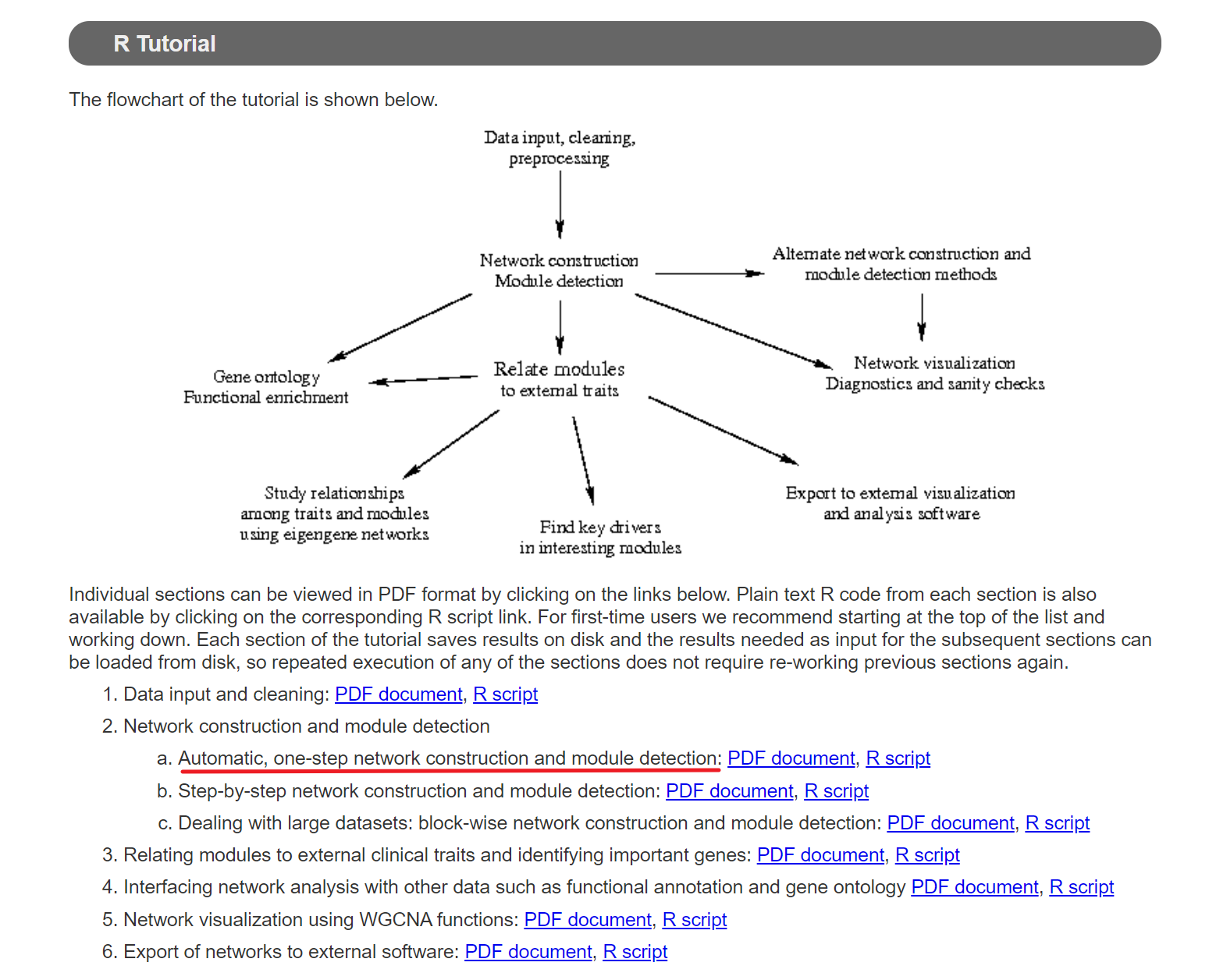
数据准备
依据前面的探索表达值筛选和cpm处理.,我们使用方案四进行后续分析
# load a test data
fs=list.files(path = 'data/project/',
pattern='PRJ',full.names = T,recursive = T)
f=fs[1]
load(f)
# get group info
pro=gsub('.Rdata','',basename(f))
library(fs)
key <- gsub(".Rdata","",basename(f),perl = TRUE)
test <- dir_info("data/all_for_wgcna", recurse = TRUE)
patho <- grep(key,test$path)
path <- test[patho,]$path
gd <- read.delim(path)
group <- as.factor(gd$infection_status)
# filter with filterByExpr
keep.exprs <- edgeR::filterByExpr(cg_exp, group=group)
cg_exp2 <- cg_exp[keep.exprs,] # 重新计算文库大小
ct <- cg_exp2
# cpm with log
datExpr4 <- edgeR::cpm(ct,log=T)
datExpr4 <- t(datExpr4)WGCNA 设置
这一步涉及到使用WGCNA包,在Rstudio中运行WGCNA时,是无法激活WGCNA的并行加速度,官方教程中有细致的说明 Tutorials for WGCNA R package, 但是这个教程很久没更新了,在查找软阈值这一步,根据CPU的状态判断,Rstudio中设置后还是可以并行的
# Load the WGCNA package
library(WGCNA)Warning: 程辑包'WGCNA'是用R版本4.0.5 来建造的载入需要的程辑包:dynamicTreeCutWarning: 程辑包'dynamicTreeCut'是用R版本4.0.3 来建造的载入需要的程辑包:fastclusterWarning: 程辑包'fastcluster'是用R版本4.0.5 来建造的
载入程辑包:'fastcluster'The following object is masked from 'package:stats':
hclustWarning: 'which'存在多个方法表
载入程辑包:'WGCNA'The following object is masked from 'package:stats':
cor# The following setting is important, do not omit.
options(stringsAsFactors = FALSE);
library(WGCNA)
# Allow multi-threading within WGCNA. This helps speed up certain calculations.
# At present this call is necessary for the code to work.
# Any error here may be ignored but you may want to update WGCNA if you see one.
# Caution: skip this line if you run RStudio or other third-party R environments.
# See note above.
enableWGCNAThreads()Allowing parallel execution with up to 7 working processes.检测缺失值
经过之前的处理,这一步一般都是没问题的 ,默认省略
# gsg = goodSamplesGenes(datExpr4, verbose = 3)
# gsg$allOK一步选择软阈值
这是教程中的模板代码,几乎通用
# Choose a set of soft-thresholding powers
powers = c(c(1:10), seq(from = 12, to=20, by=2))
# Call the network topology analysis function
sft = pickSoftThreshold(datExpr4, powerVector = powers, verbose = 5)pickSoftThreshold: will use block size 3454.
pickSoftThreshold: calculating connectivity for given powers...
..working on genes 1 through 3454 of 12952
..working on genes 3455 through 6908 of 12952
..working on genes 6909 through 10362 of 12952
..working on genes 10363 through 12952 of 12952
Power SFT.R.sq slope truncated.R.sq mean.k. median.k. max.k.
1 1 0.3730 1.720 0.985 3320.00 3230.000 5270.0
2 2 0.0554 -0.302 0.919 1280.00 1160.000 2960.0
3 3 0.5700 -1.040 0.930 606.00 498.000 1910.0
4 4 0.7570 -1.420 0.933 324.00 237.000 1330.0
5 5 0.8270 -1.570 0.949 189.00 122.000 980.0
6 6 0.8550 -1.640 0.955 117.00 66.200 747.0
7 7 0.8520 -1.700 0.949 76.50 37.800 584.0
8 8 0.8480 -1.750 0.948 51.80 22.400 467.0
9 9 0.8570 -1.760 0.955 36.30 13.800 380.0
10 10 0.8430 -1.790 0.948 26.10 8.660 313.0
11 12 0.8550 -1.790 0.961 14.30 3.620 221.0
12 14 0.8360 -1.830 0.958 8.42 1.610 161.0
13 16 0.8560 -1.800 0.974 5.22 0.763 120.0
14 18 0.8620 -1.800 0.980 3.36 0.377 92.0
15 20 0.8530 -1.820 0.982 2.25 0.193 71.7# Plot the results:
sizeGrWindow(9, 5)
par(mfrow = c(1,2));
cex1 = 0.85;
# Scale-free topology fit index as a function of the soft-thresholding power
plot(sft$fitIndices[,1], -sign(sft$fitIndices[,3])*sft$fitIndices[,2],
xlab="Soft Threshold (power)",ylab="Scale Free Topology Model Fit,signed R^2",type="n",
main = paste("Scale independence"));
text(sft$fitIndices[,1], -sign(sft$fitIndices[,3])*sft$fitIndices[,2],
labels=powers,cex=cex1,col="red");
# this line corresponds to using an R^2 cut-off of h
abline(h=0.90,col="red")
# Mean connectivity as a function of the soft-thresholding power
plot(sft$fitIndices[,1], sft$fitIndices[,5],
xlab="Soft Threshold (power)",ylab="Mean Connectivity", type="n",
main = paste("Mean connectivity"))
text(sft$fitIndices[,1], sft$fitIndices[,5], labels=powers, cex=cex1,col="red")pickSoftThreshold函数中的RsquaredCut 参数是最小无标度拓扑拟合指数 R^2,也就是纵轴,数值越高,网络越符合无标度特征 (non-scale),达到这个值的最小的power将会被选中,也有可能一直达不到,默认是0.85.
我们可以这样提取出来
sft$powerEstimate[1] 6如果一直达不到怎么办呢?这个问题在WGCNA的FAQ里有提到,详情可以去看看WGCNA package: Frequently Asked Questions (ucla.edu) 结论如下
| Number of samples | Unsigned and signed hybrid networks | Signed networks |
|---|---|---|
| Less than 20 | 9 | 18 |
| 20-30 | 8 | 16 |
| 30-40 | 7 | 14 |
| more than 40 | 6 | 12 |
sessionInfo()R version 4.0.2 (2020-06-22)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19042)
Matrix products: default
locale:
[1] LC_COLLATE=Chinese (Simplified)_China.936
[2] LC_CTYPE=Chinese (Simplified)_China.936
[3] LC_MONETARY=Chinese (Simplified)_China.936
[4] LC_NUMERIC=C
[5] LC_TIME=Chinese (Simplified)_China.936
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] WGCNA_1.70-3 fastcluster_1.2.3 dynamicTreeCut_1.63-1
[4] fs_1.5.0 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Biobase_2.50.0 sass_0.4.0 edgeR_3.30.3
[4] bit64_4.0.5 jsonlite_1.7.2 splines_4.0.2
[7] foreach_1.5.1 bslib_0.2.5.1 Formula_1.2-4
[10] assertthat_0.2.1 stats4_4.0.2 latticeExtra_0.6-29
[13] blob_1.2.1 impute_1.62.0 yaml_2.2.1
[16] backports_1.2.1 pillar_1.6.1 RSQLite_2.2.7
[19] lattice_0.20-41 glue_1.4.2 limma_3.46.0
[22] digest_0.6.27 checkmate_2.0.0 RColorBrewer_1.1-2
[25] promises_1.2.0.1 colorspace_2.0-1 preprocessCore_1.50.0
[28] htmltools_0.5.1.1 httpuv_1.6.1 Matrix_1.3-4
[31] pkgconfig_2.0.3 purrr_0.3.4 GO.db_3.11.4
[34] scales_1.1.1 whisker_0.4 jpeg_0.1-8.1
[37] later_1.2.0 htmlTable_2.2.1 git2r_0.28.0
[40] tibble_3.1.2 generics_0.1.0 IRanges_2.22.2
[43] ggplot2_3.3.5 ellipsis_0.3.2 cachem_1.0.5
[46] nnet_7.3-14 BiocGenerics_0.36.0 survival_3.2-11
[49] magrittr_2.0.1 crayon_1.4.1 memoise_2.0.0
[52] evaluate_0.14 fansi_0.5.0 doParallel_1.0.16
[55] foreign_0.8-80 data.table_1.14.0 tools_4.0.2
[58] lifecycle_1.0.0 matrixStats_0.59.0 stringr_1.4.0
[61] S4Vectors_0.26.1 munsell_0.5.0 locfit_1.5-9.4
[64] cluster_2.1.0 AnnotationDbi_1.50.3 compiler_4.0.2
[67] jquerylib_0.1.4 rlang_0.4.11 grid_4.0.2
[70] rstudioapi_0.13 iterators_1.0.13 htmlwidgets_1.5.3
[73] base64enc_0.1-3 rmarkdown_2.9 gtable_0.3.0
[76] codetools_0.2-16 DBI_1.1.1 R6_2.5.0
[79] gridExtra_2.3 knitr_1.33 dplyr_1.0.7
[82] fastmap_1.1.0 bit_4.0.4 utf8_1.2.1
[85] Hmisc_4.5-0 rprojroot_2.0.2 stringi_1.5.3
[88] parallel_4.0.2 Rcpp_1.0.6 rpart_4.1-15
[91] vctrs_0.3.8 png_0.1-7 tidyselect_1.1.1
[94] xfun_0.24