Please be aware that the RNAseqStat2 https://github.com/xiayh17/RNAseqStat2 is an new version of RNAseqStat. RNAseqStat won’t be updated any more.
The goal of RNAseqStat is a workflow for DEG analysis.

RNAseqStat
Installation
You can install the released version of RNAseqStat from CRAN with:
install.packages("RNAseqStat")And the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("xiayh17/RNAseqStat")Example
This is a basic example which shows you how to solve a common problem:
library(RNAseqStat)
#>
## basic example coderun all in one time
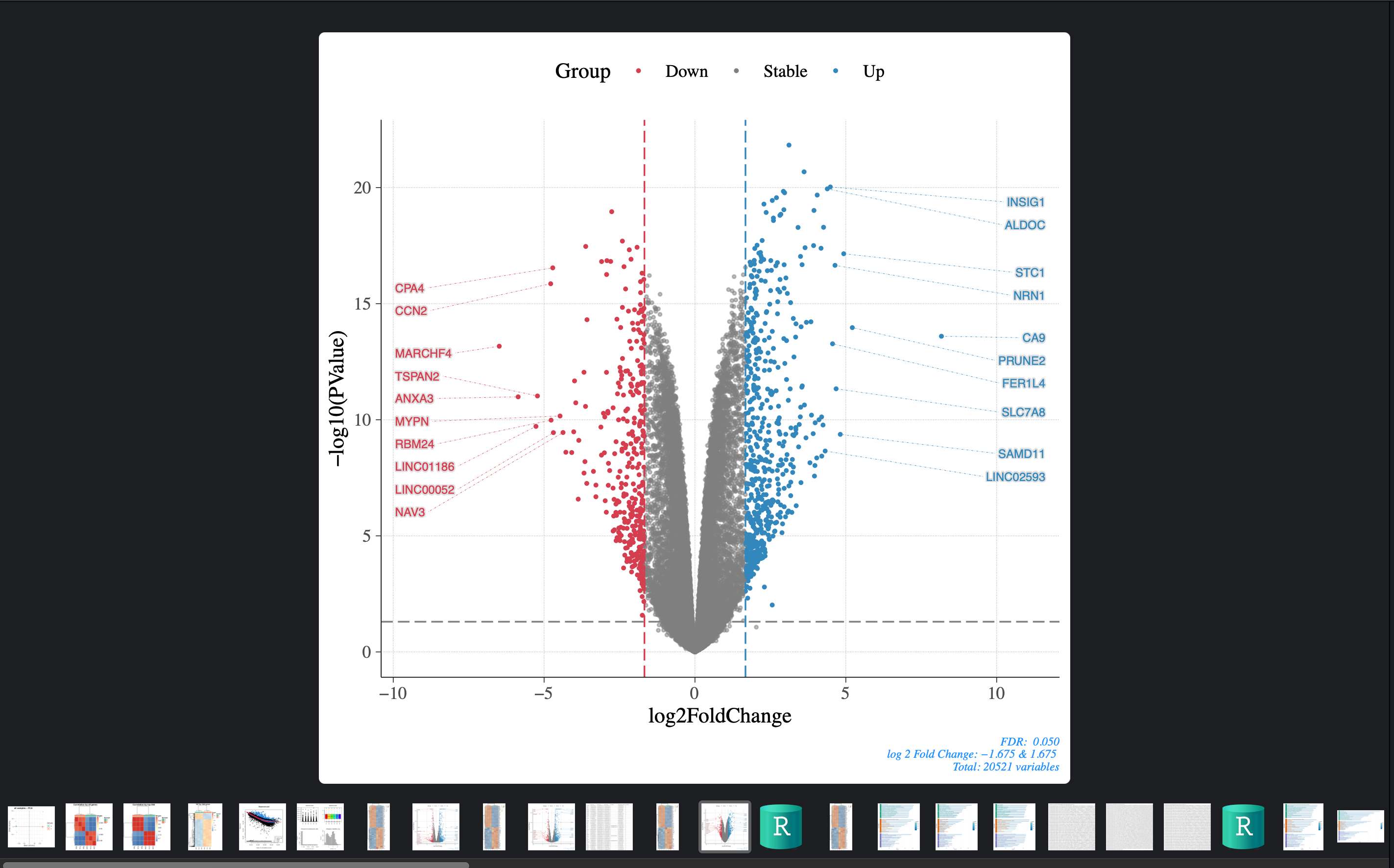
runAll will run full workflow in default parameters.
This include 5 steps:
- Check your data
- DEG analysis
- GO
- KEGG
- GSEA
runAll(count_data = counts_input, group_list = group_list,
test_group = "T", control_group = "C",
OrgDb = 'org.Hs.eg.db', dir = results_dir)